|
Differential expression analysis and detection of novel splicing or fusions events is an important focus for researchers that frequently rely on samples that have been formalin-fixed and paraffin-embedded (FFPE). Agilent introduces the new SureSelectXT RNA Direct protocol for RNA-Seq capture that, combined with world class capture libraries, enables even FFPE samples to perform reliably and generates RNA profiling data to reveal the biological state of the sample. SureSelectXT RNA Direct provides you with the ability to:
-
Generate concordant gene expression data regardless of tissue storage method [Fresh frozen (FF) and FFPE]
-
Analyze degraded FFPE RNA samples with DV200 values as low as 20%
-
Eliminate ribosomal depletion or poly(A)-enrichment and associated biases
|
|
|
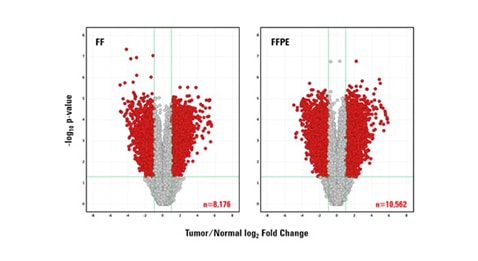
Figure 1. Concordant Tumor/Normal Expression Analysis for FF and FFPE samples. Volcano plot of significant fold-change where red points represent genes with >2-fold-change expression with corrected p-value <0.05.
|
Try the SureSelectXT RNA Direct product today!
|
|
|

Figure 2. Agilent SureSelectXT RNA Direct Protocol: Simplified RNA-Seq capture protocol that starts with total RNA and incorporates a single hybridization step and on-bead PCR post-capture to maximize the performance of the workflow.
|
|
|
 |
|

To learn more, download this Application Note today!
|
|