Access Agilent eNewsletter June 2015
>> Update My Profile | Subscribe to Access Agilent | Article Directory

Integrated ‘omics’ approach is advancing toxicology testing
By Natàlia Garcia-Reyero
Environmental Laboratory, US Army Engineer Research and Development Center Institute for Genomics Biocomputing and Biotechnology, Mississippi State University
and Mary T. McBride
Agilent Director, Government Relations
Ecotoxicology is a multidisciplinary field that integrates ecology and toxicology to study the effects of toxic chemicals on biological organisms, especially at the population or ecosystem level. Ecotoxicologists often struggle to make sense of health changes in animals because they must base their work on known mechanisms of toxicity. However, the ‘omics’ technologies (genomics, transcriptomics, proteomics, metabolomics) are changing the way researchers can approach ecotoxicology. These discovery tools provide valuable new insights, leading to improved understanding of chemical modes of action and cellular responses to stressors. We can also link cellular perturbations caused by chemical exposures to traditional adverse apical outcomes. While every ‘omics’ technology yields important information, integrating these different types of data using a systems toxicology approach can offer significantly enriched biological context, and help researchers make sense of very complex biological systems. In this article, we review results of a recent collaboration using an integrated ‘omics’ approach.
 Enlarge
Enlarge
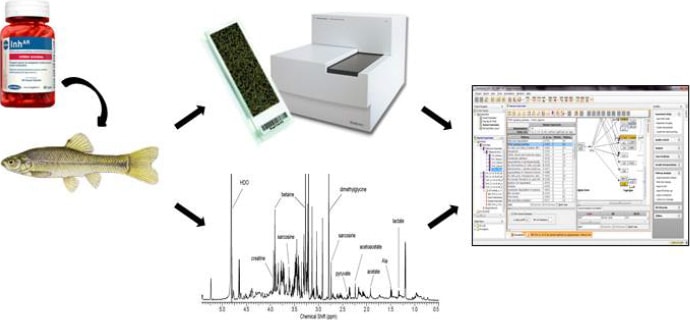
Figure 1. Fathead minnows exposed to fadrozole over various time/concentration courses. Gene expression and NMR-based metabolomics profiles were obtained for each time/dose point, and the data sets were integrated and analyzed using Agilent GeneSpring GX software.
Search, discover, and map pathways by integrating ‘omics’
Aromatase, a member of the cytochrome P450 superfamily, is an enzyme that plays a pivotal role in the conversion of testosterone to estradiol in vertebrates. Several studies have shown the potential adverse effects of aromatase inhibition on reproduction and sex ratios in birds, reptiles, and fish.[1] Garcia-Reyero and her collaborators employed an integrated systems toxicology approach that combined transcriptomics, metabolomics, and network inference. To understand the mechanistic basis of aromatase inhibition (Figure 1), [2] the team exposed fathead minnows to fadrozole (a model aromatase inhibitor) in water, and then allowed the fish to recover in clean water. The approach allowed for examination of changes caused by the adaptation and recovery processes – and infer connections at the transcript and metabolite levels. Samples were collected at various times over the course of the experiment.
An Agilent custom 15,000-feature microarray was used for gene expression analysis.[3] Normalized data were imported into Agilent GeneSpring GX bioinformatics software to identify differentially expressed genes (DEGs). The DEGs were detected over the course of the experiment (ranging from 39-112 genes per time point). Two genes were common to all exposure time points, while 116 genes (roughly a third of all DEGs) were common between the exposure and recovery phases. Functional enrichment analysis of the DEGs revealed 34 affected pathways; five pathways were common among the exposure data sets, but there were no common pathways for the recovery data.
Metabolomics aspects were captured using an Agilent Inova 600 MHz 1H NMR. Spectral analysis identified several metabolites impacted by fadrozole exposure. These included metabolites associated with the energetic capacity of the ovary, amino acid synthesis/protein degradation, and osmolytes.
Integrated systems toxicology enables deep mechanistic understanding
To understand the interplay between the various endpoints, transcriptomics and metabolomics data sets were integrated and examined for correlations. Gene expression was linked to higher-level functional effects of fadrozole by examining the mutual information correlation between gene expression data and metabolite profiles in nondirected networks. The integrated analysis revealed changes in gene expression consistent with increased testosterone in the ovaries of fathead minnows exposed to fadrozole. In addition, metabolites such as glycogen and taurine strongly correlated with increased testosterone levels. These data, combined with both in vivo and ex vivo steroidogenesis data suggest that steroidogenic enzymes, including aromatase, accumulate as a mechanism to compensate for aromatase inhibition.
New systems toxicology approaches lead the way
Our ability to measure ever-increasing numbers of biological endpoints simultaneously, combined with our improving scientific understanding of how genes, proteins, and small molecules interact at the molecular, cellular, and tissue levels, form the basis for a revolution in the way toxicity testing and risk assessments are done. Nonetheless, there are significant challenges to overcome so that the ecotoxicology community can capitalize on this ability and use increased understanding to inform risk assessments. Credible links must be established between responses measured at the molecular or cellular levels and the adverse apical responses observed in the whole organism or population level. However, systems toxicology approaches are heralding a promising new approach to ecotoxicology.
Agilent provides multiomics solutions
Integrated biology solutions from Agilent address various challenges in disease research and toxicology. With analytical solutions across genomics, transcriptomics, proteomics, and metabolomics, and the latest version of our GeneSpring GX bioinformatics software designed for pathway-centric multiomic data integration, Agilent is committed to helping you begin your own journey of integrated multiomics discovery.
References
- G.T. Ankley, et al. Toxicol. Sci., 2002, 67, 121-130.
- N. Garcia-Reyero, et al. Gen. Comp. Endocrinol., 2014, 203, 193-202.
- P. Larkin, et. al. Environ. Toxicol. Chem., 2007, 26, 1497-1506.
For Research Use Only. Not for use in diagnostic procedures.
>> Update My Profile | Subscribe to Access Agilent | Article Directory