Access Agilent eNewsletter, April 2014
>> Update My Profile | Subscribe to Access Agilent | Article Directory
Exploring top-down exposomics
By Stephen M. Rappaport
University of California, Berkeley
and Anthony Macherone
Agilent Senior Applications Chemist
Visiting Scientist, Johns Hopkins University
The exposome represents the sum of all chemical exposures received by an individual over a lifetime, from exogenous sources (food, pollutants, ionizing radiation, drugs, lifestyle factors, infections) and endogenous sources (human and microbiota metabolism, oxidative stress, lipid peroxidation, infections, and pre-existing disease). “As such, the exposome is everything that is ‘not the genome’ and serves as an umbrella for all of the traditional omes.” [1].
Exposure science is now moving away from a single point, bottom-up strategy of measuring one exposure and one effect to a top-down discovery approach. Non-targeted techniques identify relevant biomarkers, followed by targeted approaches to differentiate exposure and disease biomarkers. Measuring exposures – or, if you will, the exposome – has historically relied on self-administered questionnaires that have not changed much over the last century. However, this lack of lab-based exposomic techniques and methods is rapidly being addressed.
Measuring the exposome in blood
It is an enormous technical challenge to achieve the necessary combination of extreme multiplexing, sensitivity, and throughput for exposome-wide association studies (EWAS) and follow-up studies. We need to conduct untargeted EWAS with 10 to 50 µL of blood, serum, or plasma from each of a few hundred subjects, and to perform follow-up biomarker targeted analyses in equivalent volumes from thousands of subjects. An appropriate system comprises an LC or GC with a time-of-flight (TOF) mass spectrometer, with excellent sensitivity and mass accuracy. Follow-up investigations of promising biomarkers employs robotics and triple-quadrupole MS with multiple-reaction monitoring.
Analytical strategies
Dedicated nanoflow LC/TOF and LC/triple quadrupole, and GC/TOF, GC/triple quadrupole, are essential to ensure the necessary sensitivity and precision for data-driven EWAS and targeted, high-throughput follow-up work.
A recent untargeted analysis [2] measured more than 30,000 unique features from a single 100 µL sample of serum or plasma using reversed-phase LC and hydrophilic interaction chromatography, in positive and negative electrospray modes. An Agilent 1200 Infinity Series with an Agilent 6538 UHD Accurate-Mass Q-TOF LC/MS, ideal for a small-molecule EWAS, was used.
Another tactic modifies the Agilent-Fiehn GC/MS method [3] with an Agilent 7890B GC and Agilent 7200 Q-TOF to investigate serum extracts derivatized with methoxyamine hydrochloride and N-methyl-N-(trimethylsilyl) trifluoroacetamide.
From the general to the specific
Once biologically relevant markers are annotated and validated, targeted high-throughput analyses are required, to measure biomarkers of causal exposures and disease progression. Measuring endocrine disruptors, using the Agilent 7890B GC with an Agilent 7000C Series Triple Quadrupole, is just one example [4].
Once data are collected, sophisticated bioinformatic software is required to compare biomarker levels across populations and to investigate covariates. Agilent Mass Profiler Professional (MPP) Software provides advanced statistical analysis and visualization tools for GC/MS, LC/MS, CE/MS, and ICP-MS data analysis that can be used to identify key features and perform global analyses (Figure 1).
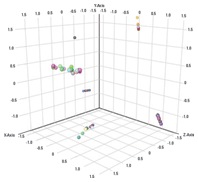 Enlarge
Enlarge
Figure 1. Principal component analysis of urinary organic aciduria biomarkers generated by Agilent Mass Profiler Professional Software.
Measuring the urinary exposome
In Maple Syrup Urine Disease (MSUD), three amino acids critical for protein synthesis are not properly metabolized and, therefore, build up to toxic levels in the blood, causing aciduria. Although most organic acidurias are directly correlated to genetic anomalies, many are also precipitated by exogenous and iatrogenic sources, such as xylene isomers (commonly found in paint solvents), and gastrointestinal dysbiosis.
Single quadrupole GC/MS identifies and quantifies urinary organic acidurias (UOA) such as MSUD, to establish a profile of the UOA exposome at any life stage. The Agilent J&W HP-5ms Ultra Inert GC column is ideal for this analysis. Baseline data can be collected at birth and compared to later data to monitor disease onset and progression. Large cohort studies of individual exposomes can be evaluated and compared to determine new UOA biomarkers using statistical analysis (Figure 1). In addition, monitoring chemical exposure can facilitate the discovery of risk factors and identify new biomarkers for most of the diseases responsible for human morbidity and mortality. This model can further help us understand the genome × environment relationship in disease.
Agilent exposomic solutions
It is estimated that 70% to 90% of chronic human diseases are caused by environmental factors that cannot be attributed to inherited genetic variability. Agilent has the tools to help you investigate this huge and largely unexplored territory, and gain new insights into many longstanding environmental threats to human and animal health. Further details are available in Agilent application notes 5991-3418EN Using the Blood Exposome to Discover Causes of Disease and 5991-3541EN Targeted Exposomics: Profiling Urinary Organic Acids.
References
- J. Pleil. J. Toxicol. Environ. Health, Part B. 15, 264 (2012).
- J. Ivanisevic et al. Anal. Chem. 85, 6876 (2013).
- M. Palazoglu, O. Fiehn. Metabolite Identification in Blood Plasma Using GC/MS and the Agilent Fiehn GC/MS Metabolomics RTL Library. Application note, Agilent Technologies, Inc. Publication number 5990-3638EN (2009).
- A. Macherone, M. Churley. Monitoring steroidal analogues in clinical and environmental chemistry: one model for exposomics. The 61st Conference on Mass Spectrometry and Allied Topics, June 9–13. Minneapolis Convention Center, Minneapolis, Minnesota (2013).
>> Update My Profile | Subscribe to Access Agilent | Article Directory
