Cancer Research: Cell Proliferation and Colony Formation Assays
January 2022
Aberrant cell proliferation rates and the propensity of individual cells to survive and propagate over long periods of time are two hallmarks of cancer. Cell proliferation assays are designed to measure the former, while the colony formation assay assesses the latter. Both proliferation and colony formation assays play a key role in cancer research and rely on accurate determination of cell number within a culture, however they differ substantially in a few important aspects. In this TekTalk we focus on how these two assays can be utilized, providing a brief description of the assay principles, and guidance on selecting the right assay format.
Cell proliferation assays
Cell proliferation assays quantify the rate in which a population of cells increase in number over time after initial cell seeding in a microplate or other culture vessel. A variety of endpoint and kinetic cell proliferation assay formats have been developed, relying on either direct or indirect readouts of cell number. Endpoint cell proliferation assays provide a single snapshot in time of cell number after a predefined experiment duration that is compared across treatment conditions. In contrast, kinetic cell proliferation assays provide a detailed profile of changes in cell number over time for each treatment condition, with cell number captured at optimized intervals, and the ability to normalize results to account for variations in starting cell number. Proliferation assays that provide indirect readouts for cell number include homogenous assays measuring total DNA or ATP levels, among many other approaches. Proliferation assays that provide a direct readout of cell number utilize either imaging or flow cytometry techniques to detect and quantify total cell number within a sample or within a representative subpopulation. Assay platforms have been developed to measure cell proliferation in both 2- and 3-D cell culture formats.
Colony formation assays
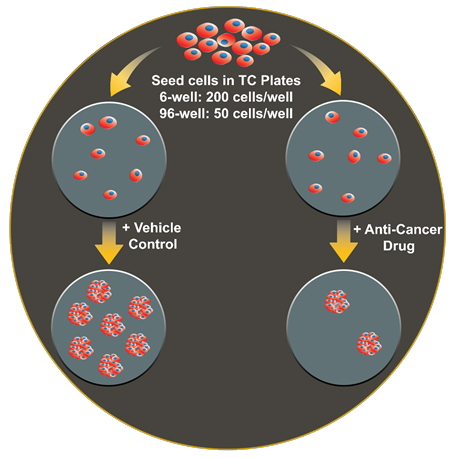
The colony formation assay, or clonogenic assay, is designed to measure the ability of a single adherent cell to survive over time and expand into a clonal population. For applications such as cancer drug screening, it is important to distinguish cells that retain this proliferative capacity from those that do not. The colony formation assay was first used to evaluate the effect of radiation on cancer cell survival and is still the preferred method for evaluating cell reproductive death after treatment with ionizing radiation [1,2]. The assay is performed by seeding cells in a microplate at a low enough density such that individual cells can propagate to a sufficient colony area without impinging on a neighboring colony. At a desired time point, usually several days to multiple weeks after seeding and/or treatment, adherent colonies are fixed then stained to enable visual inspection of the culture vessel and quantification of the number of colonies that formed. A major drawback of the colony forming assay is that scoring colonies is typically carried out manually by a trained technician. This analytical approach is both labor-intensive and limited in its ability to be carried out in a high-throughput fashion using a format larger than 12- or 24-well plates. Furthermore, although the accepted criteria for what constitutes a colony is 50+ cells, a quantitative method to assign colony size cutoffs is not frequently adhered to, leaving such manually assessed cut-offs to be subjective. Variations on the colony formation assay include the soft agar colony formation assay, which is used to measure the ability of transformed cells to grow in an anchorage-independent manner.
Conclusions
When employed successfully, proliferation assays and the colony formation assay provide valuable information about the characteristics of transformed cells. While often one or the other assay type is selected for inclusion in a given study, together they provide complementary data and greater overall insight into cancer cell behavior.
Featured Applications
Automated Kinetic Imaging Assay of Cell Proliferation in 384-Well Format
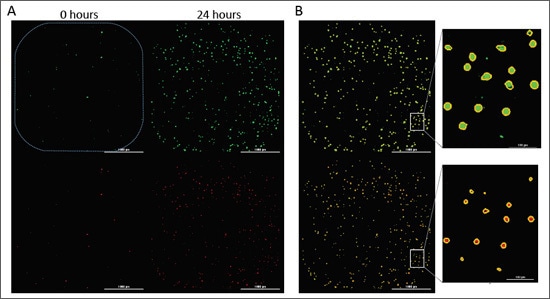
Evasion of cell death is a hallmark of cancer cells that allows them to overcome endogenous barriers to cancer development, as well as to resist treatment. Understanding how cancer cells are able to evade cell death promises to unlock new avenues to treat human cancer. Here we present a fully automated image-based assay to monitor both the proliferation and cell death of a fibrosarcoma cancer cell line in response to multiple anti-neoplastic drugs in a high-throughput format.
Automated Colony Formation Assay
The colony formation assay evaluates the proliferative capacity of a single cell. For applications such as cancer drug screening, it is important to distinguish cells that retain this proliferative capacity from those that do not. Conventional analysis of this assay involves scoring and quantifying colonies in each well of a multi-well format manually by eye, limiting its throughput capabilities. We present an automated method for conducting and analyzing the colony formation assay in both a 6-well and 96-well microplate using the upright color brightfield imaging capabilities of the Cytation 7 Cell Imaging Multimode Reader with a wide field of view.
Product Spotlight
BioTek BioSpa Live Cell Analysis System
The BioTek BioSpa live cell analysis system offers unique benefits for a wide variety of live cell imaging applications in up to 8 microplates or other labware. With 5 imaging modes, the system enables a wide range of microscopy applications.
Tek Tips
Selecting the right cell proliferation assay
Kinetic proliferation assays based on direct cell counting techniques provide a straightforward approach for generating detailed profiles of the number of cells within a population over time. Label-free direct cell counting techniques are a convenient and effective alternative to methods that rely on fluorescent labels or indirect biochemical readouts of cell population size. The High Contrast (HC) cell counting application, available with BioTek Lionheart FX Automated Live Cell Imager and Cytation Cell Imager Multimode Readers, uses modified high contrast brightfield to conduct accurate label-free cell counts.
Key advantages of kinetic cell proliferation assay using label free high-contrast brightfield:
- Convenient. Accuracy and sensitivity of direct cell counting without the limitations associated with fluorescent labels
- Robust. Automated image analysis produces results comparable to nuclear stains across wide range of cell densities
- Insightful. Detailed brightfield images provide phenotypic characterization of cell morphology and viability
Furthermore, combining label-free cell counting and fluorescent imaging of apoptosis and necrosis markers provides an accurate method for measuring treatment-induced effects on cell proliferation concurrently with normalized cell death over time.
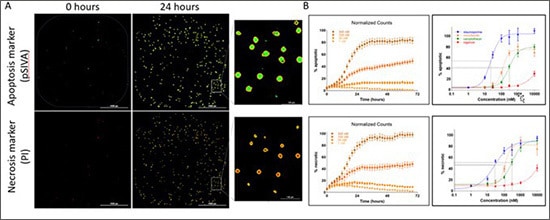
Fluorescence-based kinetic cell death analysis. (A) Example images of cells positive for pSIVA apoptosis marker and PI necrosis marker at two time. Positive apoptotic and necrotic cells are identified using size and signal threshold parameters with primary masks applied (inset shows enlarged region for clarity). (B) Kinetic profiles of % apoptotic and % necrotic are used to generate dose-response curves and EC50 values.
Considerations for optimizing the colony formation assay
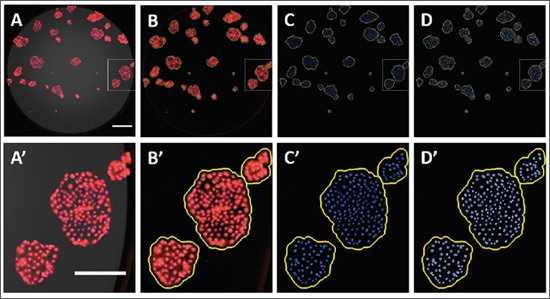
a. Calibration of colony area based on cell number: An important criterion for qualifying a cluster of cells as a colony is the presence of 50 or more cells, which is typically determined subjectively via stereoscope or by assessing directly by eye. Alternatively, by first determining the relationship between cell number and colony area, area measurements can provide a reliable metric for automated imaging-based approaches for conducting the colony formation assay. We previously described an automated approach to quantify colonies based on the fluorescence properties of Crystal Violet while verifying the number of cells using the spot counting module in the DAPI secondary mask[3]. In the example below, Caco2 and HeLa colonies stained with Crystal Violet and Hoechst 33342 were imaged with fluorescence microscopy. From this data set, a linear regression was fit to correlate colony area with their respective number of cells. The resulting linear equation enabled an estimation of the area for a 50-cell colony, which in turn was used to calibrate the cellular analysis workflow where the subpopulation of colonies containing at least 50 cells can be quantified.
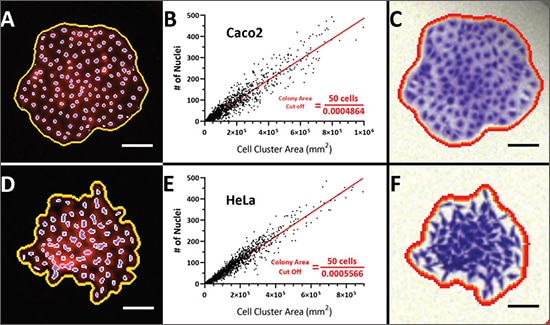
Correlation of colony size with cell number. A population of Crystal Violet and Hoechst 33342-stained Caco2 (A) and HeLa (D) cell clusters were plotted and a linear regression was then fit to correlate colony area with cell number (nuclei) (B and E). The derived linear equation was reformatted to calculate the estimated area for a 50- cell colony as indicated. A 50-cell colony for Caco2 and HeLa cells corresponds to an area of 1.03x106 and 9.0x105 mm2, respectively. These values were applied to color brightfield images of Crystal Violet-stained colonies (C and F). Scale bar = 200 μm.
b. Format: The colony formation assay can be adapted for a single dish assay (10 cm, 60 mm or 35 mm culture dish), or microplate ranging from 6-well to 96-well. The format chosen will largely depend on the number of conditions that are needed to be tested and compared. A simple +/- experiment set up can easily be done in a lower throughput format, such as a 6-well or single dish. A higher density format like a 96-well plate opens the possibly to do array comparison, such as multi-point, multi-drug dose responses. In a larger format, more colonies can be cultured and counted within the vessel, whereas high density, lower culture area microplates require a much lower initial seeding density and may be more technically challenging to reliably keep colonies spaced well enough to employ automated image analysis.
c. Cell type: Not all cell types form colonies in the same way. Epithelial cells (like Caco2) tend to want to stay attached and make for easy analysis. Others like MDA-MB-231 cells tend to want to disperse and other less straightforward assays like the soft agar assay may be more appropriate. The ability of your cell type of interest to form colonies should be empirically determined.
d. Plating Efficiency (% of cells that form colonies): Not all cells that are seeded will form colonies, and so the number of colonies will likely be less than the number of initial cells seeded. This can be influenced by the particular cell type, age or number of passages, or culture conditions and should also be empirically determined. Knowing the plating efficiency will influence the seeding density that should be performed.
Developmental Biology Resources
Webinars
Using Automated Imaging to Quantitatively Analyze the Colony Formation Assay
Presenter: Dr. Ernest Heimsath, Principal Scientist

In this webinar, we discuss automated methods for conducting and analyzing the colony formation assay using both the colorimetric and fluorescent properties of crystal violet, the well-established dye for this assay. We will cover important considerations for setting up the colony formation assay in multi-well formats, as well as discuss approaches for constructing a quantitative image analysis workflow that reliably identify colonies and extract meaningful results.
Tools and Techniques for Optimizing Cell Proliferation Studies
Presenter: Joe Clayton, Senior Principal Scientist

In this webinar we provide a detailed review of available methods for measuring cell proliferation, with a particular focus on recent assay innovations, including both 2D and 3D formats. This information will assist researchers in selecting the most appropriate and effective system for measuring cell proliferation based on a broad range of factors. Additionally, we cover important considerations for setting up and running cell proliferation assays, including techniques for improving the accuracy and reproducibility of results.
For Research Use Only. Not for use in diagnostic procedures.