Access Agilent eNewsletter June 2016

Agilent multi-omics solutions unravel the effects of low-level gamma radiation on rice seeds
S. A. Deepak, Agilent Application Development Scientist
Ionizing radiation—such as gamma radiation—severely affects growth and development of living organisms. [1] In March 2011, the Great Tohoku Earthquake in Fukushima and the tsunami that followed set off meltdowns at the Fukushima Daiichi Nuclear Power Plant (FDNPP). The meltdowns forced Japan to impose restrictions on consumption of agricultural goods produced in some parts of this area during 2012-13 because radionuclide contamination was observed. [2,3] In this article, we describe the changes that occurred in rice seeds exposed to chronic low-level gamma radiation from contaminated soil. Our study took advantage of the data integration and insights offered by Agilent multi-omics solutions (Figure 1).
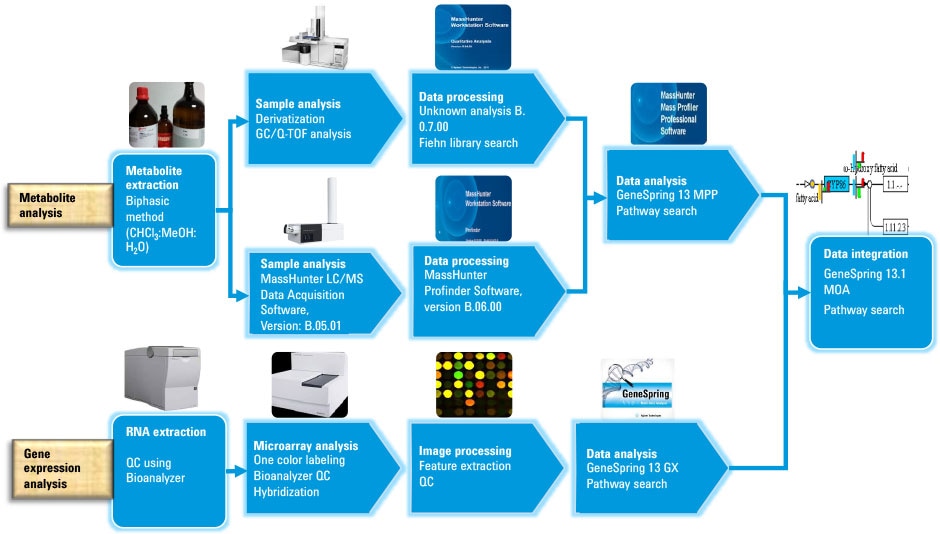
Figure 1. Effective workflow for gene expression and metabolite analysis followed by multi-omics data integration for identification of biomarkers of radiation stress in rice.
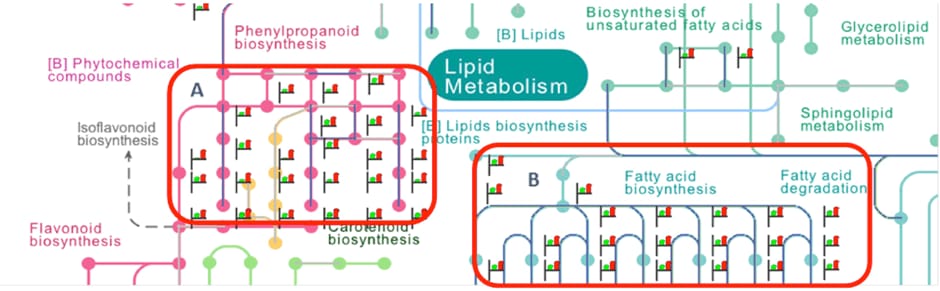
Figure 2. Differentially expressed metabolic pathways in rice due to exposure to low-level gamma radiation in rice seeds. A) Phenylpropanoid biosynthesis pathway. B) Fatty acid metabolism pathways.
Rice plants (Oryza sativa L. cultivar Koshihikari) grown in radionuclide-contaminated soil of a rice field in Iitate farm in Fukushima after the FDNPP nuclear disaster were collected and the radiation levels from soil were measured with a Germanium semiconductor detector. The radiation in harvested seeds was also measured and found to be below the allowed limits (100 bq). The seeds from the same variety of rice plants grown in Minamisoma served as control.
Identifying genes that elicit defense-related pathways
Global gene expression profiling with Agilent rice catalog microarrays followed by differential and statistical analysis with Agilent GeneSpring software revealed that 2,331 differential genes were responsive to low-level gamma radiation in rice. These genes belonged to plant defense, cell wall synthesis, secondary metabolite production, fatty acid metabolism, antioxidant, and energy cycling pathways. Most of the genes that belonged to secondary metabolism (phenylpropanoid) and fatty acid metabolism pathways were prominently up-regulated (Figure 2).
The phenylpropanoid pathway produces monolignols that serve as precursors for lignin reinforcement in the cell wall. With their partial aromatic nature, lignins are also known to resist gamma radiation. [4] The PR10 pathogenesis-related gene showed elevated expression and this gene was also identified as a potential marker for radiation stress in rice leaves exposed to radionuclide-contaminated soil from the exclusion zone around the Chernobyl reactor site. In general, a positive relationship was also observed in the expression of upstream and downstream pathway genes and metabolites.
LC/MS and GC/MS provide comprehensive coverage of metabolites
With the complementary analytical techniques LC/MS and GC/MS, we enhanced the coverage of metabolites and identified a total of 383 metabolites from rice seeds. Differential analysis that used the Agilent Mass Profiler Professional (MPP) module of GeneSpring identified fifty metabolites in rice seeds exposed to low-level gamma radiation. These differential metabolites belonged to energy, fatty acid, amino acid, nucleotide, and secondary metabolic pathways in rice. The amino acid proline, an important component of cell wall proteins, was highly up-regulated, with a +17.5-fold change in radiated seeds. With an important role in the protection of the cell against free radicals induced by harmful environmental effects, its accumulation is a good indicator of stress tolerance in plants.
Pathway-centric multi-omic data integration reveals plant defense mechanisms
The Multi-Omic Analysis (MOA) module of GeneSpring/MPP 13.1 was used to combine the gene expression and metabolome data. The results showed overlapping entities in several pathways between the two -omics (Table 1). Two metabolites, linolenic acid and 12-oxo-phytodienoic acid (12-OPDA), along with the ACX gene in alpha-linolenic acid metabolism, were up-regulated. This pathway is involved in the production of the hormone jasmonic acid, which is associated with stress responses of plants. A positive correlation in the regulation of the genes and metabolites was evident, which revealed a well-coordinated defense against radiation in rice seeds. These results suggest that a multi-omics approach provides better understanding of biological complexity.
| Pathway | Gene expression | Metabolites | ||
|---|---|---|---|---|
| Matched entities | Pathway entities | Matched entities | Pathway entities | |
| Purine metabolism | 3 | 132 | 2 | 92 |
| Pentose phosphate pathway | 2 | 48 | 1 | 35 |
| Sulfur metabolism | 1 | 35 | 1 | 29 |
| Cysteine and methionine metabolism | 4 | 85 | 3 | 57 |
| alpha-Linolenic acid metabolism | 3 | 33 | 1 | 40 |
| Pyrimidine metabolism | 2 | 112 | 1 | 66 |
| Glycine, serine and threonine metabolism | 3 | 55 | 2 | 51 |
| Sphingolipid metabolism | 2 | 22 | 1 | 25 |
| Aminoacyl-tRNA biosynthesis | 2 | 114 | 4 | 52 |
| Arginine and proline metabolism | 2 | 63 | 3 | 90 |
Table 1. Some overlapping entities in rice pathways between gene expression and metabolite data.
Agilent integrated biology solutions
To help scientists find meaningful interpretations of complex biological data, Agilent provides instrumentation and software to perform multi-omic research that includes genomics, transcriptomics, proteomics, and metabolomics. Agilent GeneSpring 13.1 bioinformatics software accelerates pathway-centric multi-omic data integration.
For all the details of this study, explore the complete Application Note 5991-6416EN to learn more about how the Agilent GeneSpring multi-omics workflow was used to perform gene expression and metabolite analysis on samples derived from plant sources. Researchers demonstrated that integrated -omic analysis that used a combination of Agilent platforms and software tools was a powerful approach to identify the major events that occur in plants that undergo stress. Agilent wishes to thank all authors of that Application Note, which made this article possible.
References
- G. Hayashi et al. “Unraveling low-level gamma radiation-responsive changes in expression of early and late genes in leaves of rice seedlings at Iitate village, Fukushima”. J. Hered. 2014, 723-738.
- M. Saito. “Fukushima rice passes radiation tests for first time since disaster”. Reuters, January 4, 2015.
- T. Imanaka et al. “Early radiation survey of Iitate village, which was heavily contaminated by Fukushima Daiichi accident, conducted on 28 and 29 March 2011”. The Radiation Safety J, 2012, 680-686.
- F. E. Brauns and D. A. Brauns. “The chemistry of lignin: Covering the literature for the years 1949—1958”. Academic Press Inc. New York, USA, 1960, 564.
Stay informed about the applications that are important to you
Subscribe to Access Agilent
Our free customized
monthly eNewsletter
All articles in this issue
-
 New Agilent 8900 ICP-QQQ delivers increased sensitivity, improved accuracy, and higher productivity
New Agilent 8900 ICP-QQQ delivers increased sensitivity, improved accuracy, and higher productivity -
 New Agilent 1260 Infinity II LC system delivers higher level of operational efficiency
New Agilent 1260 Infinity II LC system delivers higher level of operational efficiency -
 Robust Agilent 6470 Triple Quadrupole LC/MS delivers confident quantification and streamlined workflows
Robust Agilent 6470 Triple Quadrupole LC/MS delivers confident quantification and streamlined workflows -
 Achieve accurate analysis of sulfur and nitrogen with the new and simplified Agilent 8355 SCD
Achieve accurate analysis of sulfur and nitrogen with the new and simplified Agilent 8355 SCD -
 Agilent 1290 LC and 6550 iFunnel Q-TOF combine to provide fast, high-resolution peptide mapping of innovator and biosimilar mAbs
Agilent 1290 LC and 6550 iFunnel Q-TOF combine to provide fast, high-resolution peptide mapping of innovator and biosimilar mAbs -
 Agilent collaborates with Academia—from investigating arsenic mysteries to understanding the complexity of biology
Agilent collaborates with Academia—from investigating arsenic mysteries to understanding the complexity of biology -
 Excellent inertness for analysis of challenging polar compounds
Excellent inertness for analysis of challenging polar compounds -
 Detection of semivolatile extractables and leachables (E&Ls) found in pharmaceutical products
Detection of semivolatile extractables and leachables (E&Ls) found in pharmaceutical products -
 Agilent multi-omics solutions unravel the effects of low-level gamma radiation on rice seeds
Agilent multi-omics solutions unravel the effects of low-level gamma radiation on rice seeds -
 Agilent comprehensive water screening solution reliably identifies wastewater contaminants of emerging concern:
Agilent comprehensive water screening solution reliably identifies wastewater contaminants of emerging concern:
A real-life application
Figure 1

Effective workflow for gene expression and metabolite analysis followed by multi-omics data integration for identification of biomarkers of radiation stress in rice.
Figure 2

Differentially expressed metabolic pathways in rice due to exposure to low-level gamma radiation in rice seeds. A) Phenylpropanoid biosynthesis pathway. B) Fatty acid metabolism pathways.