Agilent Innovations Enable Biologists to "See" DNA and RNA Inside Single Cells
|
|
April 29, 2013Biology researchers can visualize segments of DNA on a chromosome or RNA in individual cells at high resolution because of innovations at Agilent Laboratories and Agilent's Genomics Business.
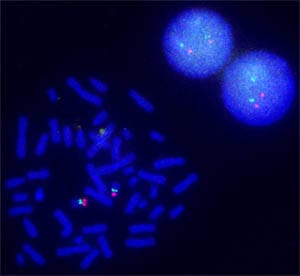
Agilent researchers dramatically simplified and advanced a procedure whose origins go back to the 1960s. They improved precision and removed all limits on the types of genomes that can be studied by FISH. The oFISH technique exposes chromosomes or cells to a pool of oligonucleotides that have fluorescent molecules attached to them. These fluorescently decorated probes bind to the complementary strands of DNA or RNA wherever they reside in a person's cells. When a probe binds to its target, its fluorescent tag provides a way for researchers to see its location.
Visualizing and mapping genetic material in an individual cell are important for understanding a variety of chromosomal abnormalities and other genetic mutations. Changes in DNA can cause cells to make a protein that doesn't work as it should. Health problems could result if DNA is duplicated, if it moves from one chromosome to another, or if a chunk of DNA is flipped or rearranged. Agilent's oFISH Advantage Previously, scientists had to make FISH probes from bacterial sources that had human DNA inserted in their bacterial chromosomes. Because these bacterial artificial chromosomes (BACs) are much larger than the changes in DNA that can impact health, scientists could not visualize important small-scale changes. Libraries of BAC probes are also cumbersome to generate and maintain, subject to contamination and laborious to purify, therefore limiting FISH to situations for which BACs were readily available. Agilent fabricates very high quality oligonucleotides using a technique developed from inkjet technology to "print" sequences of guanine, adenine, thymine, and cytosine, the building blocks of DNA recorded using the letters G, A, T, and C. Combined with this ability to print DNA, Agilent designs oFISH probes on a computer and therefore manufactures the probes reproducibly by chemical synthesis. Researchers simply order Agilent SureFISH probes online. Users now have a powerful tool to analyze what previously were very difficult regions for FISH in any situation of interest – such as those too small or too near complicated DNA structures for BAC FISH probes to be available. In addition oFISH can be done for organisms for which BAC probes have not been made. Agilent's advances make oFISH research available for much broader use for any species, including bacteria. University Research Collaborations A number of collaborations enabled Agilent researchers to verify the key advantages of Agilent's oFISH compared to traditional BAC technology, including the following and recent joint publications: The University of Arizona -- Arthur R. Brothman, Ph.D., FACMG, Professor of Pathology,
University of California, San Francisco -- Shinya Yamanaka, MD, Ph.D., Senior Investigator at the Gladstone Institute of Cardiovascular Disease, L.K. Whittier Foundation Investigator in Stem Cell Biology, Professor of Anatomy and winner of the 2012 Nobel Prize in Physiology or Medicine for his discovery of how to transform ordinary adult skin cells into cells that, like embryonic stem cells, can then develop into other cell types.
2012 Barney Oliver Prize for Innovation Agilent Laboratories awarded the scientists who developed oFISH the 2012 Barney Oliver Prize for Innovation. The Prize honors Bernard M. Oliver (1916 – 1995), who was a scientist, inventor, and innovator. Agilent Laboratories has awarded the prize annually since 1999 for contributions to Agilent that result from work done in the Labs and that demonstrate Barney's outstanding qualities of creativity, innovation, technical depth, breadth of expertise, and respect for business value. |